Revista Científica UDO Agrícola Volumen 9.
Número 4. Año 2009. Páginas: 770-775
Phenotypic and
genotypic screening of rice genotypes at seedling stage for salt tolerance
Selección fenotípica y genotípica de genotipos de arroz para tolerancia a
la salinidad en la etapa de plántulas
Soubir TITOV1, Salil KUMAR BHOWMIK ![]() 2, Mirza
MOFAZZAL ISLAM3, Ayesha SIDDIKA2, Sharmin
SULTANA2 and Md. SHAHIDUL HAQUE2
2, Mirza
MOFAZZAL ISLAM3, Ayesha SIDDIKA2, Sharmin
SULTANA2 and Md. SHAHIDUL HAQUE2
1Biotechnology and Genetic Engineering
Discipline, Khulna University, Khulna-9208, Bangladesh; 2Department
of Biotechnology, Bangladesh Agricultural University, Mymensingh-2202,
Bangladesh and 3Bangladesh Institute of Nuclear Agriculture, P. O.
Box 4, Mymensingh-2200, Bangladesh
E-mail:
salil_kist@yahoo.com ![]() Corresponding author
Corresponding author
|
Received: 08/03/2009 |
First reviewing ending:
10/12/2009 |
|
First review received: 11/27/2009 |
Accepted: 28/12/2009 |
ABSTRACT
Selection
for salinity tolerant genotypes of rice based on phenotypic performance alone
is less reliable and will delay in progress in breeding. Recent advent of
molecular markers, microsatellites or simple sequence repeats (SSRs), have been
useful in finding salt tolerant rice genotypes. Three selected SSR markers
already known to be polymorphic, viz.,
RM7075, RM336 and RM253, were used to evaluate rice genotypes for salt
tolerance. Phenotypic and genotypic evaluation for salinity tolerance was done
at the seedling stage. Phenotyping was done in
hydroponic system using salinized (EC 12 dS/m)
nutrient solution following IRRI standard protocol. Large variation in salinity
tolerance among the rice germplasms was detected.
Salt stress (EC 12 dS/m) reduced seedling height by
19.0% and total dry matter of tolerant lines by 40.6%, whereas, total dry
matter of susceptible lines were reduced by 46.0-73.5%. All the tested markers
were polymorphic and were able to discriminate salt tolerant genotypes from
susceptible. The genotypes having similar banding pattern with Pokkali were considered as salt tolerant. Markers RM7075,
RM336 and RM253 identified eight, nine and seven salt tolerant genotypes,
respectively. Through phenotypic and genotypic study, three genotypes viz., Pokkali,
TNDB-100 and THDB were identified as salt tolerant rice genotypes. These SSR
markers might have sequence homology with salt tolerant rice genotypes and
consequently the markers could able to identify salt tolerant rice genotypes
from susceptible ones.
Key words: rice, salinity
tolerance, SSR markers, seedling stage.
RESUMEN
La
selección para resistencia a la salinidad de genotipos de arroz, basada
solamente en el comportamiento fenotípico, es menos confiable y retarda el avance
en el mejoramiento. Se han utilizado avances recientes en marcadores
moleculares, microsatélites o repeticiones de
secuencias simples (SSR por sus siglas en inglés) para determinar genotipos de
arroz tolerantes a la salinidad. Se utilizaron tres marcadores SSR viz., RM7075,
RM336 y RM253 para evaluar genotipos de arroz para tolerancia a la salinidad.
La evaluación fenotípica y genotípica para la tolerancia a la salinidad se
realizó en la etapa de plántula. La fenotipificación
de once genotipos se realizó en un sistema hidropónico utilizando solución
nutritiva salinizada (CE 12 dS/m). Se siguió el
protocolo estandarizado del IRRI para evaluar la tolerancia a la salinidad. Se
detectó una gran variación en la tolerancia a la salinidad entre el germoplasma
de arroz. La altura de las plántulas y la materia seca total de las líneas
tolerantes se redujeron en un 19,0 y 40,6%, respectivamente, bajo estrés salino
(CE 12 dS/m), en tanto que las de las líneas
susceptibles se redujeron en un 46,0% y 73,5%, respectivamente. Los marcadores
mostraron polimorfismo y fueron capaces de discriminar los genotipos tolerantes
a la salinidad de aquellos susceptibles. Los genotipos con un patrón similar de
bandas a Pokkali se consideraron como tolerantes a la
salinidad. Los marcadores SSR (RM7075, RM336 y RM253) identificaron ocho, nueve
y siete genotipos tolerantes a la salinidad, respectivamente. A través del
estudio fenotípico y genotípico, tres genotipos viz., Pokkali,
TNDB-100 y THDB se identificaron como cultivares de arroz tolerantes a la
salinidad. Estos marcadores SSR podrían tener homología de secuencias con
genotipos de arroz tolerantes a la salinidad y por consiguiente, los marcadores
podrían ser capaces de identificar genotipos de arroz tolerantes a la salinidad
de aquellos susceptibles.
Palabras clave:
Arroz, tolerancia a la salinidad, marcadores SSR, etapa de plántulas.
INTRODUCTION
Rice is the staple
food of more than 50% of the world’s population (Aggarwal
et al., 2002). By the year 2025, 21%
increase in rice production will be needed over the production of 2000 (Bhuiyan et al.,
(2002). Salinity is one of the major constraints to productivity in rice
growing areas worldwide, which is an ever-present threat to crop yield.
Therefore, development of salt tolerant varieties has been considered as one of
the strategies to increase rice production in salinity-prone coastal areas. The
response of rice to salinity varies with growth stage. Several studies have
indicated that rice at early seedling stage (2-3 leaf stage) and during
pollination and fertilization is more sensitive to salinity than during,
germination or vegetative growth stage or late reproductive stages (IRRI,
1967).
Screening of germplasm at seedling stage is readily acceptable as it is
based on a simple criterion of selection; whereas rapid screening becomes
difficult at vegetative and reproductive stages (Gregorio et al., 1997). Screening under controlled condition has the benefit
of reduced environment effects and the hydroponic system is free from complex
variations associated with soil related stress factors. The conventional
methods of plant selection for salt tolerance are not easy because of the large
effects of the environment and low narrow sense heritability of salt tolerance
(Gregorio, 1997). This hinders the development of an accurate, rapid and
reliable screening technique. However, DNA markers seem to be the best
candidates for efficient evaluation and selection of plant material. Recent
progress and technical advances in DNA marker technology permits rapid and
improved accuracy in breeding for traits prone to pronounced environmental effects
leading to poor selection efficiency.
SSR or
microsatellite markers have been proved to be ideal for making genetic maps
(Islam, 2004; Niones, 2004), assisting selection (Bhuiyan, 2005) and studying genetic diversity in germplasms. SSR markers play an important role while
identifying gene for salt tolerance or in introgressing
the genes to develop new cultivars. The aim of the present study was to screen
rice germplasm for salinity response and to evaluate
microsatellite markers for the identification of salt tolerant genotypes at the
seedling stage.
MATERIALS
AND METHODS
Plant
materials
Eleven rice germplasm accessions, with diverse genetic background, were
used in this study. Of which six were Bangladeshi landraces, four were
Bangladesh Institute of Nuclear Agriculture (BINA) developed mutants, and one
salt tolerant Indian variety ‘Pokkali’ was used as
check.
Phenotypic study of salinity
tolerance at seedling stage
The genotypes were screened for salt tolerance at seedling stage in
hydroponic system using IRRI standard protocol (Gregorio, 1997). Salinized and
non-salinized setups with three replications were maintained. The evaluation
was done using Yoshida et al. (1976)
nutrient solution at the glasshouse. The nutrient solution was salinized by
adding crude salt to obtain desired EC (12 dS/m). The modified standard evaluation system was used in
rating the visual symptoms of salt toxicity (IRRI, 1997). Visual rating of
salinity tolerance was done according to table 1. This scoring discriminated
the susceptible from the tolerant and the moderately tolerant genotypes.
Initial and final scoring was done at 13 days and 22 days after salinization. Other observations are seedling height, root
length and total dry matter recorded both at salinized and non-salinized
conditions.
|
Table
1. Modified standard evaluation score of visual salt injury at seedling stage
(Method adapted from Gregorio et al., (1997)). |
||
|
Score |
Observation |
Response
category |
|
1 |
Normal growth with no leaf symptoms |
Highly tolerant |
|
3 |
Nearly normal growth, but leaf tips or few
leaves whitish and rolled |
Tolerant |
|
5 |
Growth severely retarded; most leaves rolled;
only a few are elongating |
Moderately tolerant |
|
7 |
Complete cessation of growth; most leaves dry;
some plants dying |
Susceptible |
|
9 |
Almost all plants dead or dying |
Highly susceptible |
CTAB mini
preparation DNA extraction
DNA isolation was done from fresh leaf tissues
of 14-day old seedlings. DNA was extracted using the mini preparation CTAB
method. Grinding of leaf sample with
extraction buffer and SDS was followed by incubating the leaf sap at 65°C for
10 min. 100 ml NaCl and 100 ml
CTAB were added sequentially and mixed well; and incubated again at 65°C for 10
minutes. After that the suspensions were transferred to a new plate. 900 ml chloroform : isoamyl (24:1) was
added and mixed well by a shaker. The sample was then centrifuged at 5700 rpm
for 10 minutes. After that the supernatant were transferred into new eppendorf tubes. Then 600 ml
ice-cold isopropanol was added into the new eppendorf
tubes and shaken slowly and then centrifuged at 5700 rpm for 15 minutes. The
supernatant was decanted and air dried for at least one hour. Pelletes were washed with 70% ethanol (200 ml), spinned for 15 minutes at 5700 rpm and then air-dried for
1/2-1 hours. Then the ethanol was removed and air-dried. The pelletes were resuspended in 30.0
ml X TE
buffer.
Amplification
of microsatellite markers and evaluation of genotypes
Three selected primers were used for this study as those were used
previously by Islam (2004); Bonilla et al. (2002); Niones (2004) and Gregorio et al. (2002) in recombinant inbred
lines (RILs) of Pokkali X IR29 for tagging salt
tolerance genes, where Pokkali was salt tolerant and
IR29 was salt susceptible. Among them RM7075, RM336 and RM253 were polymorphic
and showed clear bands Each PCR reaction was carried out with 15.0µl reaction
mixtures containing 1.5 µl 10 X buffer, 0.75 µl dNTPs,
1µl primer forward, 1µl primer reverse, 0.5 µl taq
polymerase, 8.25 µl ddH2O and 2.0 µl of each template DNA samples.
PCR profile was maintained as initial denaturation at 94oC for 5
min, followed by 34 cycles of denaturation at 94oC for 1 min,
annealing at 55oC for 1 min and polymerization at 72oC
for 2 min; and final extension by 7 min at 72oC. Banding pattern of
the genotypes was scored comparing
the banding pattern of Pokkali. The germplasm lines that showed similar banding pattern like Pokkali,
were considered as tolerant and that with different banding pattern were
considered as susceptible.
Data
analysis
Data obtained were
subjected to one way analysis of variance for completely randomized design.
Treatment means were compared using Least Significant Difference. Correlation
coefficients of different traits at seedling stage under salinized condition
were also calculated.
RESULTS
AND DISCUSSION
Screening
of genotypes for salt tolerance at seedling stage
All
genotypes grew robust and were uniform in colour and
height in the non-salinized condition. In salinized condition, the
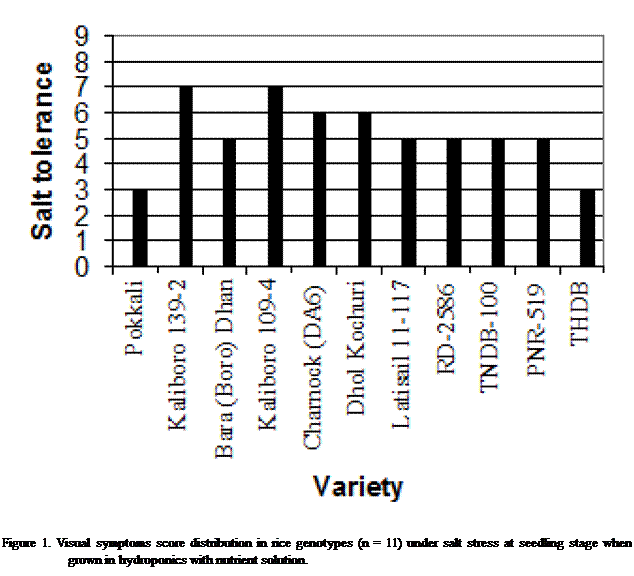
genotypes showed wide ranging variation in phenotypes from score 1 (highly
tolerant) to 9 (highly susceptible) (Figure 1). The most salinity tolerant germplasm were Pokkali, THDB, and
TNDB-100. Four moderately salinity tolerant genotypes were identified as
RD-2586, PNR-519, Dhol Kochuri
and Bara Dhan. The most susceptible salt tolerant
genotypes were Kaliboro 139-2 and Kaliboro
109-4. The modified standard evaluation system of IRRI (Gregorio et al., 1997) was used in rating the
visual symptoms of salt injury.
Seedling height was shorter in salinized condition, compared to the seedlings
grown in non-salinized conditions (Table 2). Seedling height and total dry
matter of susceptible genotypes showed higher percent reduction than tolerant
genotypes. Lower percent reduction of seedling height was recorded in genotypes
Pokkali and THDB followed by genotypes TNDB-100,
RD-2586, PNR-519 and Dhol kochuri. On the other hand, higher percent reduction of
seedling height was showed by genotypes, Kaliboro
139-2 and Kaliboro 109-4. The percent reduction of
total dry matter ranged from 40-75. Lower percent reduction of total dry matter
was found in genotypes Pokkali, TNDB-100 and THDB. In
contrast, Kaliboro 139-2 and Kaliboro
109-4 showed higher percent reduction of total dry matter. Tolerant cultivars
showed less growth reduction than sensitive genotypes under salinized
conditions (Suplick-Ploense et al., 2002).
|
Table 2. Seedling height
and total dry matter in rice genotypes (n = 11)
under both non-saline and saline conditions at seedling stage when grown in hydroponics
with nutrient solution. |
|||||||
|
Serial No. |
Genotypes |
Seedling
height (cm) |
Total dry matter (g) |
||||
|
Non-salinized |
Salinized |
% reduction |
Non-salinized |
Salinized |
% reduction |
||
|
1 |
Pokkali |
61 |
50 |
18 |
9.12 |
5.48 |
40 |
|
2 |
Kaliboro
139-2 |
62 |
34 |
45 |
9.17 |
2.54 |
72 |
|
3 |
Bara
(Boro) Dhan |
55 |
39 |
29 |
7.7 |
3.61 |
53 |
|
4 |
Kaliboro
109-4 |
59 |
31 |
47 |
11.73 |
2.97 |
75 |
|
5 |
Charnock
(DA6) |
42 |
30 |
29 |
7.25 |
3.26 |
55 |
|
6 |
Dhol Kochuri |
59 |
44 |
25 |
10.69 |
5.28 |
51 |
|
7 |
Latisail
11-117 |
55 |
40 |
27 |
8.44 |
3.75 |
56 |
|
8 |
RD-2586 |
44 |
34 |
23 |
6.7 |
3.85 |
43 |
|
9 |
TNDB-100 |
42 |
33 |
21 |
7.38 |
4.33 |
41 |
|
10 |
PNR-519 |
49 |
37 |
24 |
5.78 |
3.12 |
46 |
|
11 |
THDB |
40 |
33 |
18 |
8.47 |
4.97 |
41 |
|
LSD
(0.05) |
1.7 |
1.4 |
- |
0.442 |
0.208 |
- |
|
All the eleven genotypes
showed a wide variation in phenotypes. Salt tolerant seedlings were distinct from
the sensitive seedlings grown in salinized condition. Seedlings grown in
salinized condition showed different visual symptoms of salt injury. The
symptoms were prominent on the first and second leaves and were visualized by
leaf rolling, formation of new leaf, brownish and whitish of leaf tip, drying
of leaves and also reduction in root growth, stunted shoot growth with
thickened stem leading to a complete cessation of growth and dying of seedlings
(Gregorio et al., 1997).
Salinity in rice was associated with Na+ exclusion and increased
absorption of K+ to maintain a good Na+/K+
balance in the shoot under saline condition. It is considered that damage of
leaves was attributed to accumulation of Na+ from the root to the shoot in external
high concentration (Lin et al.,
2004). In several species including rice,
salt stress might increase or even include the expression of specific genes and
repress or completely suppress the expression of others (Hasegawa et al., 2000).
At the seedling
stage, highly significant and positive correlations were found between plant
height and total dry matter at salinized condition and correlations between
salt tolerance and plant height, total dry matter and root length were inverse
and significant (Table 3). This implies that salt tolerant genotypes (with
lower salt tolerance score) exhibited higher plant height and total dry matter.
Peng et al.
(1999) reported that increasing plant height would allow greater biomass
production. Zhang et al.
(2004) found similar result in their study with doubled haploid (DH) population
consisting of 81 lines. They reported that increase of plant height was
responsible for increase in biomass; so as to increase yield potential. It is
crucial to note that Pokkali, THDB and TNDB-100
genotypes showed higher plant height and total dry matter and also performed as
salt tolerant.
Table 3. Correlation
of different traits at seedling stage of rice under salinized condition.
|
|||
|
Traits |
Salt tolerance |
Seedling height |
Total dry matter |
|
Seedling
height |
-0.406 ** |
|
|
|
Total dry matter |
-0.740 ** |
0.622 ** |
|
|
Root length |
-0.278 * |
0.115 ns |
0.291
* |
|
*
= Significant at 5% level of probability **
= Significant at 1% level of probability ns
= Non significant at > 5% level of probability |
|||
On the basis of standard evaluation system score and
phenotypic performance, three genotypes (Pokkali,
THDB and TNDB-100) were identified as salt tolerant and RD 2586, Dhol Kochuri, PNR-519, Bara (Boro) Dhan, Latisail
11-117 and Charnock (DA6) were identified as
moderately tolerant at seedling stage.
Screening
of salt tolerance through SSR markers
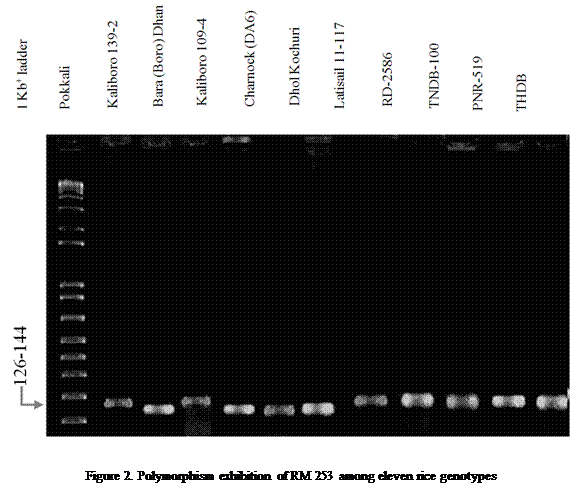
Three SSR markers,
RM7075, RM336 and RM253, were used to evaluate germplasms
for salinity tolerance. The bands obtained from other genotypes were compared
to the band obtained from Pokkali. Pokkali was used as salt tolerant genotype in this study
because it is known as salt tolerant genotype. The germplasms
having similar banding pattern to Pokkali were
considered as tolerant while others having different banding pattern to Pokkali were considered as susceptible. In case of RM253,
Bara (Boro) Dhan, Latisail 11-117, RD-2586, TNDB-100, PNR-519 and THDB were
found as tolerant. On the other hand, Kaliboro 139-2,
Kaliboro 109-4, Charnock
(DA6), and Dhol Kochuri
were found as susceptible with RM253 (Figure 2).
Considering the primer
RM7075, genotypes Charnock (DA6), Dhol
Kochuri, Latisail 11-117,
RD-2586, TNDB-100, PNR-519 and THDB were found as tolerant whereas Kaliboro 139-2, Bara (Boro) Dhan, and Kaliboro 109-4 were
found as susceptible. Bara (Boro) Dhan,
Charnock (DA6), Dhol Kochuri, Latisail 11-117,
RD-2586, TNDB-100, PNR-519 and THDB were identified as tolerant and Kaliboro 139-2 and Kaliboro 109-4
were identified as susceptible with RM336.
The marker RM7075
identified eight tolerant and three susceptible genotypes in comparison with Pokkali. Out of genotypically
identified eight salt tolerant genotypes, three were tolerant and five were
moderately tolerant based of phenotypic performance. With marker RM336, nine
genotypes exhibited as salt tolerant and two genotypes were susceptible.
Phenotypically three tolerant, six moderately tolerant at seedling stage were
identified amongst the nine genotypically tolerant
genotypes. Seven tolerant and four
susceptible cultivars were found when eleven genotypes were tested with RM253.
Considering these seven genotypically salt tolerant
genotypes, phenotypically three tolerant and four moderately tolerant at
seedling stage. Bhuiyan (2005) identified 158
tolerant individuals of the F2 and F3 population of BRRI Dhan
28 X PSBRc88 with the marker RM493. Moreover, he observed 105 tolerant
individuals phenotypically.
Considering both
phenotypic and genotypic observations, three genotypes TNDB-100, THDB and Pokkali were identified as salt tolerant. Four genotypes i.e. RD 2586, Dhol
Kochuri, PNR-519 and Bara (Boro)
Dhan were characterised as
moderately tolerant. The selected markers (RM7075, RM336 and RM253) showed good
level of polymorphism with the eleven rice genotypes. These SSR markers were
able to discriminate well tolerant genotypes from susceptible. Thus these
markers have a clear relationship with salt tolerance alleles studied in rice
genotypes. Molecular marker helps to identify alleles that are associated with
key phenotypic traits (Xu et al., 2004). Nguyen et al., (2001) found that the marker
RM315 had association with NaCl tolerant alleles at
seedling population (IR64/ChengHui 448, IR64/OM1706
and IR64/FR13A) under EC 18 dS/m and salt stress
genes were located at loci in chromosomes 1 and 8. Similar result was reported
by Lang et al., (2000). They found that RM223 was closely linked to salt
tolerance gene in chromosome 8. Since, the markers that were used in this study
showed polymorphism, these markers could be proficiently used in tagging salt
tolerant genes, in marker-assisted selection and quantitative trait loci (QTL)
mapping; and identified salt tolerant rice genotypes could be used in the
improvement of salt tolerant rice genotypes.
LITERATURE
CITED
Aggarwal, R. K.; V. V. Shenoy, J. Ramadevi, R. Rajkumar and L. Singh. 2002. Molecular
characterization of some Indian Basmati and other elite rice genotypes using
fluorescent AFLP. Theor. Appl. Gent. 105: 680-690.
Bhuiyan, M. A. R. 2005. Efficiency in evaluating salt
tolerance in rice using phenotypic and marker assisted selection. M. Sc. Dissertation, Department of Genetics and Plant Breeding,
Bangladesh Agricultural University, Mymensingh,
Bangladesh. 96
p.
Bhuiyan, N. I.; D. N. R.
Paul and M. A. Jabber. 2002. Feeding the extra millions by 2025: challenges
for rice research and extension in Bangladesh. In: Proceedings of the National Workshop on Rice Research and
Extension, Bangladesh Rice Research Institute, Gazipur,
January 29-31, 2002.
Bonilla, P. S, J. Dvorak, D. Mackill, K. Deal
and G. Gregorio. 2002. RFLP and SSLP mapping of salinity tolerance
genes in chromosome 1 of rice (Oryza saliva L.)
using recombinant inbred lines. The Philippines Agricultural Scientist 85:
64-74.
Gregorio,
G. B. 1997. Tagging salinity tolerant genes in rice using
amplified fragment length polymorphism (AFLP). Ph. D. Dissertation. University of the Philippines Los Baños
College, Laguna, Philippines. 118 p.
Gregorio, G. B.; D. Senadhira and R. D.
Mendoza. 1997. Screening rice for salinity tolerance. IRRI
Discussion Paper Series no. 22. Manila (Philippines): International Rice
Research Institute. p. 1-30.
Gregorio, G. B.; D. Senadhira, R.
D. Mendoza, N. L. Manigbas, J. P. Roxas and C. Q. Guerta. 2002. Progress in
breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Research 76: 91-101.
Hasegawa, P. M.; R. A. Bressan, J. K. Zhu and
H. J. Bonhert. 2000. Plant cellular and molecular
responses to high salinity. Annu. Rev. Plant Physio. Plant Mol.
Biol. 51: 463-499.
International
Rice Research Institute (IRRI). 1967. Annual Report for 1967. Los Baños, Laguna, Philippines. 308 p.
International
Rice Research Institute (IRRI). 1997. Rice Almanae. IRRI-WARDA-CIAT, Los Baños, Laguna,
Philippines.
Islam,
M. M. 2004. Mapping salinity tolerance genes in rice (Oryza sativa L.) at reproductive stage.
Ph.D. Dissertation, University of the Philippines Los Baños
College, Laguna, Philippines. 1-149 p.
Lang, N. T.; S. Yanagihara and B. C. Buu. 2000. Quantitative trait loci for salt tolerance in
rice via molecular markers. OmonRice 8: 37-48.
Lin, H. X.;
M. Z. Zhu, M. Yano, J. P. Gao, Z. W. Liang, W. A. Su, X. H. Hu, Z. H. Ren and
D. Y. Chao. 2004. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt
tolerance. Theor. Appl. Genet. 108: 253-260.
Nguyen, T. L.; S. Yanagihara and B. C. Buu. 2001. A microsatellite marker for a gene conferring salt
tolerance on rice at the vegetative and reproductive stages. SABRAO J. Breed. Genet. 33: 11-20.
Niones, J. M. 2004. Fine mapping of the salinity tolerance
gene on chromosome 1 of rice (Oryza sativa L.)
using near-isogenic lines. M. Sc. Dissertation, University of the
Philippines Los Baños College, Laguna, Philippines. 78 p.
Peng, S.; K. G. Cassman, S. S. Virmani, J. Sheehy
and G. S. Khush. 1999. Yield potential trends
of tropical rice since the release of IR8 and the challenge of increasing rice
yield potential. Crop Sci. 39: 1552-1559.
Suplick Ploense, M. R.; Y. L. Qian and J.
C. Read. 2002. Salinity tolerance of Texas bluegrass, Kentucky bluegrass, and
their hybrids. Crop Sci. 42: 2025-2030.
Xu, Y.; H. Beachell and S. R. McCouch. 2004. Marker based
approach to broadening the genetic base of rice in the USA. Crop Sci. 44:
1947-1959.
Yoshida,
S.; D. A. Forno, J. H. Cook and A. K. Gomez. 1976. Laboratory manual
for physiological studies of rice. International Rice Research
Institute (IRRI), Los Baños, Laguna, Philippines. p.
61-66.
Zhang,
Z. H.; P. Li, L. X. Wang, Z. L. Hu, L. H. Zhu
and Y. G. Zhu. 2004. Genetic dissection of the relationships of biomass
production and partitioning with yield and yield related traits in rice. Plant
Sci. 167: 1-8.
Página diseñada por Prof. Jesús Rafael
Méndez Natera
TABLA
DE CONTENIDO DE LA REVISTA CIENTÍFICA UDO AGRÍCOLA



