Revista Científica UDO Agrícola Volumen 7. Número 1.
Año 2007. Páginas: 15-21
Stability
analysis of safflower (Carthamus tinctorius L.) lines adaptability
in dryland conditions in Iran
Análisis de estabilidad de la adaptabilidad de líneas de cártamo (Carthamus
tinctorius L.) a condiciones de secano en Irán
Khoshnood ALIZADEH DIZAJ
Oilseed,
Food and Feed Legumes Department. Dryland Agricultural Research Institute, PO
Box 119, Maragheh, Iran. Telephone:
+98 (421) 2228078. Fax: +98
(421) 2222069. E Mail: khoshnod2000@yahoo.com
|
Received: 12/10/2006 |
First reviewing ending: 04/01/2006 |
First review received: 04/18/2006 |
|
Second reviewing ending: 05/07/2007 |
Second review received: 05/19/2007 |
Accepted: 06/04/2007 |
ABSTRACT
Spatial variability
is inherent in all field trials. The selection efficiency of the most desirable
safflower genotypes can be improved by identifying the underlying spatial
patterns in field trials and by incorporating these into the statistical
analysis. The main objective of this study was to evaluate the grain yield
stability of 25 safflower lines after adjustment for spatial variability across
a series of simple lattice designed trials at five research stations over a
three years period. There was spatial variability in 50% of the trials. For
most of the environments tested in this study, the use of complete blocks to account for variability was more efficient
than incomplete blocks. Three types of spatial analysis were effective in
accounting for variability: 1) randomized complete block design with first
order auto-correlated errors along rows, 2) lattice design with correlated
errors along rows as well as columns and 3) randomized complete block design
with first order auto-correlated errors in plots along rows and along columns.
Two genotypes (287 and 79-299) had the best stability, using the environmental
coefficient of variation. However, these were also amongst those with the
lowest yields. Yet, when the regression coefficient (b) on the basis of best linear unbiased estimates of
grain yield was used, genotypes 367
and PI250596 were the most stable. It is recommended that first a best model be
identified to describe the spatial variation in data, followed by evaluation of
the genotypes based on that model.
Key words: Carthamus
tinctorius, genotype x
environment interaction, spatial analysis.
RESUMEN
La variabilidad espacial es inherente en todos los
ensayos de campo. La eficiencia de la selección de los genotipos más deseables
del cártamo puede mejorarse identificando los patrones espaciales subyacentes
en los ensayos de campo e incorporando éstos en el análisis estadístico. El
objetivo principal de este estudio fue evaluar la estabilidad del rendimiento
de semillas de 25 líneas de cártamo después del ajuste de la variabilidad
espacial a través de una serie de ensayos diseñados en un láttice simple en
cinco estaciones de investigación en un período de tres años. Hubo una
variabilidad espacial en el 50% de los ensayos. Para la mayoría de los
ambientes evaluados en este estudio, el uso de bloques completos para explicar
la variabilidad fue más eficiente que los bloques incompletos. Tres tipos de
análisis espacial fueron efectivos para explicar la variabilidad: 1) diseño de
bloques completos al azar con errores de primer orden autocorrelacionados a lo
largo de las hileras, 2) diseño de láttice con errores correlacionados a lo
largo de las hileras así como de las columnas y 3) diseños de bloques completos
al azar con errores de primer orden autocorrelacionados a lo largo de las
hileras y de las columnas. Dos genotipos (287 y 79-299) tuvieron la mejor
estabilidad, usando el coeficiente de variación ambiental. Sin embargo, éstos
estuvieron también entre aquellos con los rendimientos más bajos. Aún, cuando
se usó el coeficiente de regresión (b) basado en las mejores estimaciones
lineales no sesgadas del rendimiento de semillas, los genotipos 367 y PI250596
fueron los más estables. Se recomienda que primero se identifique el mejor
modelo para describir la variación espacial en los datos, seguido por la
evaluación de los genotipos basada en ese modelo.
Palabras
clave: Carthamus tinctorius, interacción genotipo x ambiente,
análisis espacial.
INTRODUCTION
Development
of oilseed crops has gained a high priority in Iranian agriculture in recent
years. Drylands occupy over 6.2 million hectares of arable lands across the
country. Preliminary trials have indicated that safflower (Carthamus
tinctorius) is the oilseed crop best adapted to the low rainfall and stress
conditions of Iranian dryland (Rashid et al., 2002). In the regional crop
variety testing trials, more than 150 domestic and exotic lines of safflower
have been evaluated over eight years for grain yield in the Dryland
Agricultural Research Institute (DARI), Maragheh, Northwest Iran (Alizadeh,
2003). The relative performance of lines varies with environment, and this
genotype´environment (GxE) interaction hampers selection of lines for cultivation
over a wide region. In addition, field trials are often conducted in fields
that are quite heterogeneous due to biotic and abiotic factors, including
topography and soil fertility. The fact that crop response varies within a field,
due to underlying crop growth processes and their responses to concomitant soil
process variables in space (Nielsen et al., 1994) and time (Stafford,
1999), is a dilemma to soil and crop scientists (Cassel et al., 2000). Although experimental designs usually account for a
large section of heterogeneity in the field, a considerable amount of variation
within the blocks may remain unaccounted for by traditional methods of
analysis, especially as trial size increases as more genotypes are tested.
Modern
methods of analysis can further help to reduce this unaccounted component of
variability (Singh, 2002). The best method should have the ability to explain
data according to a standard statistical criterion. Spatial variability arises
from both variation in soil properties and distribution (i.e. natural
variation) and experimental procedure (i.e. extraneous variation) such as
effects of serpentine harvesting of plots and variation due to unequal plot
lengths arising from inaccurate trimming (Gilmour et al., 1997). An effective evaluation of cultivars can be made by
identifying and understanding both the underlying spatial pattern of
experimental material and incorporating these patterns into the statistical
analysis. Spatial analyses have been reported for cereals (Cullis and Gleeson,
1991; Grondona et al., 1996; Gilmour et al., 1997; Wilkinson et al., 1983) and pasture (Sarker et al., 2001).
Various
statistical models have been presented in the literature to study GxE
interactions (Becker, 1981; Eberhart and Russell, 1966; Finlay and Wilkinson,
1963; Kempton, 1984; Lin et al.,
1986; Plaisted and Peterson, 1959; Perkins and Jinks, 1968). The multitude of
concepts and measures of stability has been developed based on the variety of
different outlooks of experimenters and the uniqueness of their specific
problems. For example, Smith et al.
(2001) used multiplicative mixed models and adjustments for spatial field
trends, while Feyerherm et al.,
(2004) constructed statistical method for producing probabilistic inferences of
future yielding ability from a sample of cultivar performance
trials. However, the author is unaware of any reports on the use of
spatially-adjusted means for stability analysis in any crop system.
The
analyses detailed in this study were designed to (i) evaluate the spatial
variability in safflower field trials, (ii) study the adaptation of these lines
using some stability parameters on mean grain yields of safflower after
adjusting for spatial variability and (iii) suggest selections made using this
approach amongst 25 varieties from 13 field test environments.
MATERIALS AND METHODS
Twenty-five
safflower pure lines (Table 1), developed at the Dryland Agricultural Research
Institute, were evaluated over a three year period (2000 to 2003) across five
Research Stations in Iran and there were 13 growing environments in total,
because on two sites (Kurdistan and Maragheh) investigations were performed for
two years (Table 2). The individual trials were conducted using a square lattice
design with 2 replications. The experiments were planted in the late autumn of
each year just before the first frost in each region. Each genotype was sown in
plots (
Table 1. Origin of the 25
genotypes of safflower (Carthamus
tinctorius L.).
|
||
|
No. |
Genotype
|
Origin
|
|
1 |
287 |
Iran |
|
2 |
79-299 |
Iran |
|
3 |
301 |
Iran |
|
4 |
336 |
USA |
|
5 |
338 |
Syria |
|
6 |
342 |
USA |
|
7 |
348 |
USA |
|
8 |
350 |
Canada |
|
9 |
356 |
Cyprus |
|
10 |
361 |
Pakistan |
|
11 |
367 |
Kenya |
|
12 |
368 |
Spain |
|
13 |
372 |
Pakistan |
|
14 |
375 |
Pakistan |
|
15 |
376 |
Pakistan |
|
16 |
405 |
Syria |
|
17 |
406 |
Turkey |
|
18 |
411 |
Iran |
|
19 |
412 |
Iran |
|
20 |
415 |
Iran |
|
21 |
Cyprus |
Cyprus |
|
22 |
Zarghan |
Iran |
|
23 |
PI250596 |
USA |
|
24 |
PI250537 |
Canada |
|
25 |
PI258417 |
Iran |
Table 2. Location,
elevation and meteorological data for the five research sites in Iran.
|
||||||||
|
Site |
Env. |
Location |
Elevation (m) |
Year |
Prec. (mm) |
Mean Abs. Max. T (°C) |
Mean Abs. Min. T (°C) |
No. of days below 0°C |
|
Shirvan |
1 |
57° 55¢ N, 37° 23¢ E |
1086 |
2000-2001 |
186 |
17 |
3.2 |
89 |
|
2 |
2001-2002 |
329 |
20 |
1.2 |
65 |
|||
|
3 |
2002-2003 |
302 |
10.5 |
1.5 |
98 |
|||
|
Kurdistan |
4 |
47° 0¢ N, 35° 20¢ E |
1500 |
2001-2002 |
350 |
17 |
0.8 |
104 |
|
5 |
2002-2003 |
382 |
8 |
0 |
119 |
|||
|
Kermanshah |
6 |
34° 20¢ N, 47° 20¢ E |
1351 |
2000-2001 |
432 |
18 |
3.55 |
79 |
|
7 |
2001-2002 |
413 |
21 |
2 |
76 |
|||
|
8 |
2002-2003 |
424 |
14 |
1.5 |
76 |
|||
|
Ilam |
9 |
46° 25¢ N, 33° 38¢ E |
1363 |
2000-2001 |
413 |
22 |
4 |
11 |
|
10 |
2001-2002 |
627 |
23 |
5 |
13 |
|||
|
11 |
2002-2003 |
474 |
24 |
5.3 |
15 |
|||
|
Maragheh |
12 |
37° 15¢ N, 46° 20¢ E |
1720 |
2001-2002 |
381 |
18 |
1 |
114 |
|
13 |
2002-2003 |
367 |
8.5 |
0 |
134 |
|||
|
Env.: Growing environment;
Prec.: Precipitation; Mean Abs. Max. T: Mean absolute maximum temperature;
Mean Abs. Min. T: Mean absolute minimum temperature |
||||||||
Eighteen models covering a range of spatial patterns
were generated for analyzing the grain yield from each trial (Singh, 2002). The
components of spatial patterns comprised factorial combinations of block
structures, trends and structures for plot errors (Table 3).
|
Table
3. List and abbreviations of models used to describe spatial variability in
randomized complete block design (Rc) or lattice design (Lt). |
|
|
Error/ Trends |
Abbreviation |
|
Independent plot errors |
Rc
(or Lt) |
|
First
order auto-regressive errors along rows |
Rc
(or Lt)Ar |
|
First order
auto-regressive error along rows and along columns |
Rc
(or Lt)ArAr |
|
Fixed
linear trend along rows |
Rc
(or Lt)L |
|
Fixed
linear trend along rows and first order auto-regressive errors along rows |
Rc
(or Lt)LAr |
|
Fixed linear
trend along rows and first order auto-regressive error along rows and along
columns |
Rc
(or Lt)LArAr |
|
Random
cubic spline in column number (including linear trend) |
Rc
(or Lt)Cs |
|
Random
cubic spline in column number and first order auto-regressive errors along
rows |
Rc
(or Lt)CsAr |
|
Random
cubic spline in column number and first order auto-regressive error along
rows and along columns |
Rc
(or Lt)CsArAr |
Genotype effects were assumed to be fixed parameters,
while replication effects and block effects within replications
were assumed to
be random variables. Parameters
were estimated using the residual maximum likelihood (REML) method in Genstat 5
Release 4.1 (1997). The REML directive produced a statistic, called the
deviance (Dev), which facilitated the computation of the Akaike (1974)
criterion (AIC). The deviance is minus twice the REML log-likelihood ignoring a
constant depending on the fixed terms. Since the maximum
log-likelihood value is expected to increase with the number of parameters,
this criterion decreases this value by introducing a penalty in terms of the
number of unknown parameters of the variance-covariance of the error components.
Thus the AIC is based on a penalized log-likelihood, where the penalty
increases with the number of variance-covariance parameters in the fitted
spatial structure. A comparison of models
with the same set of fixed effects was carried out using the AIC. When
expressed in terms of the deviance values, this can be defined as: AICD= Dev +
2N, where N is the number of linear and non-linear variance components of the
models.
The model with the lowest AICD value was deemed to be
the best, due to goodness of fit of that model over others (Singh, 2002). The significance
of the fixed linear trend was tested using the Wald statistic (Genstat 5
committee, 1997). This is computed as the ratio of the squared estimate of the
linear trend to its estimated variance and follows a chi-square distribution in
the absence of a trend. If the trend is statistically significant at P ≤
0.05, then the best model is
chosen from models including a linear trend factor. For each trial, the
best model was used to compute the efficiency of the method of analysis This
was assessed by comparing the average variance of pair-wise genotype
comparisons with that of a randomized complete block design with independent
errors (i.e. no spatial errors) as following:

The best model was identified as describing the spatial
variation in the data. Finally, evaluations of the genotypes were made using a
combination of the spatially adjusted best model and the stability analysis
from the best linear unbiased estimates (BLUEs). The stability indices
suggested by Francis and Kannenberg (1978) (CV) and Finlay and Wilkinson (1963) (b) were calculated as following using MS
Excel.

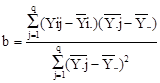
Where, Yij denotes the mean value of i-th
genotype in the j-th environment.
RESULTS
Spatial analysis of the data revealed no evidence for
the existence of fixed errors in these trials since the Wald statistics were
not significant across all environments (environment
|
Table 4. Information based on the Akaike criterion
expressed in terms of deviance values (AICD) to select the best model for
safflower trial in environment 1. |
||||
|
Model |
q |
Df |
AICD |
Walda |
|
Rc |
2 |
23 |
270.06 |
- |
|
RcAr |
3 |
22 |
268.38 |
- |
|
RcArAr |
4 |
21 |
264.71 |
- |
|
Lt |
3 |
22 |
268.30 |
- |
|
LtAr |
4 |
21 |
268.20 |
- |
|
LtArAr |
5 |
20 |
267.54 |
- |
|
RcL |
2 |
22 |
265.21 |
0.01 |
|
RcLAr |
3 |
21 |
263.26 |
0.14 |
|
RcLArAr |
4 |
20 |
259.48 |
0.01 |
|
LtL |
3 |
21 |
263.37 |
0.08 |
|
LtLAr |
4 |
20 |
262.97 |
0.31 |
|
LtLArAr |
5 |
19 |
262.05 |
0.90 |
|
RcCs |
3 |
21 |
265.21 |
0.00 |
|
RcCsAr |
4 |
20 |
263.26 |
0.14 |
|
RcCsArAr |
5 |
19 |
259.48 |
0.01 |
|
LtCs |
4 |
20 |
263.37 |
0.08 |
|
LtCsAr |
5 |
19 |
262.94 |
0.31 |
|
LtCsArAr |
6 |
18 |
262.46 |
0.92 |
|
Abbreviations used for spatial models are
defined in Table 3. q: number of variance components in the model. Df: residual degrees of freedom. aWald
statistics for testing for a linear trend along rows. |
||||
|
Table 5. Best models, efficiency over randomized
complete block design in thirteen safflower trials in dryland condition. |
||
|
Environment No. |
Best
model |
Efficiency
(%) |
|
1 |
RcArAr |
177 |
|
2 |
RcAr |
127 |
|
3 |
Rc |
100 |
|
4 |
Rc |
100 |
|
5 |
RcArAr |
148 |
|
6 |
LtArAr |
451 |
|
7 |
Rc |
100 |
|
8 |
Rc |
100 |
|
9 |
RcArAr |
82 |
|
10 |
Rc |
100 |
|
11 |
RcAr |
98 |
|
12 |
Rc |
100 |
|
13 |
Rc |
100 |
|
Environments are defined in Table 2. Abbreviations used for spatial models are
defined in Table 3. |
||
Unadjusted
means along with average best linear
unbiased estimates of grain yield over the environments and some
stability parameters including coefficient of variation (CV), regression
coefficient (b) and coefficients of determination (r2) are presented
in Table 6. The unadjusted mean
genotype grain yield over all environments ranged from 851 kg/ha to 1271 kg/ha,
whereas the observed range for adjusted means was 920-1411 kg/ha and 12 lines
had mean grain yield above the grand mean (1137 kg/ha). To demonstrate
interrelationship of the stability statistics estimated, correlation
coefficient between genotype ranks obtained from used stability indices and
mean grain yield were calculated (Table 7). A significant positive rank
correlation was obtained between genotype means, b and r2.
|
Table 6. Average
safflower grain yield (kg/ha) in all environments (Mean) along with mean best
linear unbiased estimates (BLUE) and estimates of common stability indices. |
|||||
|
Genotypes No. |
Unadjusted Mean |
BLUE |
CV |
b |
r2 |
|
1 |
907 |
923 |
0.77 |
0.60** |
0.90 |
|
2 |
958 |
1028 |
0.88 |
0.80** |
0.99 |
|
3 |
1120 |
1204 |
1.13 |
1.19* |
0.95 |
|
4 |
1114 |
1217 |
1.17 |
1.21 |
0.91 |
|
5 |
1116 |
1191 |
1.03 |
1.04 |
0.90 |
|
6 |
1271 |
1411 |
1.16 |
1.42** |
0.94 |
|
7 |
939 |
1013 |
0.95 |
0.78 |
0.81 |
|
8 |
957 |
1043 |
0.88 |
0.78* |
0.90 |
|
9 |
912 |
1015 |
0.99 |
0.85 |
0.89 |
|
10 |
936 |
1037 |
0.88 |
0.79** |
0.94 |
|
11 |
1141 |
1219 |
0.94 |
1.01 |
0.96 |
|
12 |
1116 |
1213 |
1.14 |
1.22** |
0.98 |
|
13 |
1073 |
1211 |
1.26 |
1.26 |
0.86 |
|
14 |
905 |
977 |
0.92 |
0.75* |
0.87 |
|
15 |
1028 |
1107 |
0.96 |
0.93 |
0.97 |
|
16 |
1163 |
1245 |
1.09 |
1.21** |
0.98 |
|
17 |
1025 |
1114 |
1.17 |
1.15* |
0.98 |
|
18 |
945 |
1040 |
1.00 |
0.90 |
0.95 |
|
19 |
851 |
920 |
0.93 |
0.74** |
0.93 |
|
20 |
1002 |
1109 |
1.07 |
0.97 |
0.84 |
|
21 |
1212 |
1326 |
0.95 |
1.10 |
0.95 |
|
22 |
1197 |
1311 |
1.09 |
1.27** |
0.99 |
|
23 |
1097 |
1218 |
0.99 |
1.07 |
0.98 |
|
24 |
982 |
1053 |
0.98 |
0.86 |
0.88 |
|
25 |
1190 |
1288 |
1.01 |
1.13 |
0.94 |
|
LSD 5% |
112 |
123 |
|
|
|
|
Genotypes are defined in Table 1. CV:
Coefficient of variation b: linear response to changes in
environments r2:
Coefficient of determination. * Significantly different from 1.0 at the P ≤ 0.05. ** Significantly different from 1.0 at the P ≤ 0.01. LSD:
Least Significant Difference |
|||||
|
Table 7. Correlation between
genotype ranks on the basis of mean grain yield (Mean) and stability indices. |
|||
|
|
Mean |
CV |
b |
|
CV |
0.60 ** |
|
|
|
b |
0.88 ** |
0.88 ** |
|
|
r2 |
0.44 * |
0.13 |
0.41 * |
|
CV: Coefficient of
variation. b: linear response to
changes in environments. r2: Coefficient
of determination. * and **
Significant at P ≤ 0.05 and
P ≤ 0.01, respectively. |
|||
DISCUSSION
In all but one of the trials, the use of complete blocks
to account for variability had higher efficiency than incomplete blocks (Table
5). Irrespective of the specific form of the model, and acknowledging that the
spatial variability of each field is unique (Gilmour et al., 1997), because of relatively high numbers of genotypes,
lattice design was expected to be more efficient. However, the models based on
complete blocks and first order auto-regressive errors were frequently found to
give an improvement in our field trials during these years.
There was no evidence of fixed linear trend along rows
or random cubic splines in columns. It may be concluded that natural variation,
which may result in ‘linear trend’ according to Gilmour et al. (1997), could be well described by blocking in our
experiments. Meanwhile, if a larger number of trials were examined, the
situation may change and other patterns of spatial variability might become
evident. Sarker et al. (2001)
reported all spatial variability models in 53 lentil trials. Since the selected
models accounted most effectively for spatial variability, they would therefore
enhance the breeding efficiency in the selection of the desired genotypes.
Wide
adaptation is important for safflower in dryland conditions, because of the
wide range of environments encountered. Environmental coefficient of variation
(CV), as Type 1 stability index (Lin et
al., 1986), may be considered relevant for this purpose. A highly
significant positive rank correlation was obtained between CV and mean grain
yield indicating that lower CVs were accompanied by lower grain yields (Table
7). This was expected according to Becker (1981). Although wide adaptation may
be desirable, it is difficult to achieve in practice (Becker, 1981). In terms
of CV, genotypes 287 and 79-299 were amongst those with the highest stability
(lowest CV), but they were amongst those which produced the lowest yields
(Table 6). On the other hand, Lin et al.
(1986) noted that when variability in response can be satisfactorily expressed
by a regression model, the regression coefficient (b) can serve as a stability
parameter and could be preferred to other parameters. The values of the
coefficients of determination (r2) from individual linear regression
analysis ranged from 0.81 to 0.99 (Table 6). Hence the regressions accounted
for quite a large amount of the variation across environments. However, it
should be denoted that the regression is partly auto correlated and the slope
is very much determined by the yield in the high yielding environments.
Furthermore, the regression coefficient
provides information on the shape of response along with its variation. Linear
responses to changes in environments (b) ranged from 0.6 to 1.42 (Table 6). The
large variation in regression coefficients indicates that some of the 25
entries responded differently to varying environmental conditions. Seven
genotypes showed average stability (i.e. regression coefficients did not differ
significantly from 1.0) with the grain yield above the grand mean, indicating
that they have general adaptability (Table 6). Amongst these seven entries,
genotypes 367 and PI250596 had some of the lowest CV values (Table 6) which
were selected for use in on-farm trials for demonstration.
CONCLUSIONS AND RECOMMENDATIONS
Two genotypes
(287 and 79-299) had the best stability with rather low grain yield which may
be suitable for marginal lands. When the regression coefficient (b) on the basis of best linear unbiased estimates of
grain yield was used, genotypes 367
and PI250596 were the most stable lines for dryland conditions. Regarding
efficiencies of best models over the randomized complete block design and since
the criterion used was based on maximum information in the data and a penalty
function, the inferences drawn from the best model could give most realistic
assessment of the stability of genotypes. Hence, it is recommended that to
evaluate safflower genotypes first a best model be identified to describe the
spatial variation in the data, and then the evaluation of the genotypes should
be made using it.
ACKNOWLEDGMENT
I would like to thank M. Singh from the International
Center for Agricultural Research in the Dry Areas (ICARDA) for his kind advice
and help in statistical analysis. The collaborations of M. Eskandari, H.
Hatamzadeh, A. Shariati and M. P. Siahbidi at different Research Stations of
Iran are greatly acknowledged.
LITERATURE CITED
Akaike, H.
Alizadeh, Kh. 2003. Oilseed crops for cold drylands of Iran. In:
Proceeding of 7th International conference on development of dry
lands. Tehran,
Iran. p. 33-34.
Becker, H. C. 1981. Correlations among some statistical
measures of phenotypic stability. Euphytica 30: 835-840.
Cassel, D. K., O. Wendroth and D. R. Nielsen.
2000. Assessing spatial variability in an agricultural experiment station
field. Agron. J. 92: 706-714.
Cullis, B. R. and A. C. Gleeson, 1991. Spatial
analysis of field experiments - an extension to two dimensions. Biometrics 47:
1449-1460.
Eberhart, S. A. and W. A. Russell. 1966. Stability
parameters for comparing varieties. Crop Sci. 6: 36-40.
Feyerherm, A.; G. Paulsen and A. Fritz.
Finlay, K. W. and G. N. Wilkinson, 1963. The
analysis of adaptation in a plant breeding programme. Australian J. of Agric.
Res. 14: 742-754.
Francis, T. R. and L. W.
Kannenberg. 1978. Yield stability studies in short-season maize.
Genstat 5 Committee. 1997. Genstat 5 Release 4.1,
Reference Manual Supplement, Lawes.
Gilmour, A. R.; B. R. Cullis and
A. P. Verbyla. 1997. Accounting for natural and extraneous variation in the
analysis of field experiments. J. of Agric., Bio. and Env. Stat. 2: 269-293.
Grondona, M. O.; J., Crossa; P. N. Fox and W. H. Pfeiffer. 1996.
Analysis of variety yield trials using two-dimensional separable ARIMA
processes. Biometrics, 52: 763-770.
Kempton, R. 1984. The use of biplots in
interpreting variety by environment interactions. J. of Agric. Sci. 103:
123-135.
Lin, C. S.; M. R. Binns and L. P. Lefkovitch.
1986. Stability Analysis: where do we stand? Crop Sci. 26: 894-900.
Nielsen, D. R.; O. Wendroth and
M. B. Parlange. 1994. Developing site-specific technologies for sustaining
agriculture and our environment. In: Narayanasamy, G. (Ed.).Management of land
and water resources for sustaining agriculture and our environment. Indian
Society of Soil Science, New Delhi. p. 42-47.
Perkins, J. M. and J. L. Jinks. 1968.
Environmental and genotype-environment components of variability. Heredity 23:
339-356.
Plaisted, R. L. and L. C. Peterson.
Rashid, A.; A. Beg; A. A. Attary; S. S. Pourdad and Kh. Alizadeh. 2002. Oilseed
crops for the highlands of CWANA. ICARDA Caravan 16:27-29.
Sarker, A.; M. Singh and W. Erskine. 2001.
Efficiency of spatial methods in yield trials in lentil (Lens culinaris ssp. culinaris).
J. of Agric. Sci. 25: 312-319.
Singh, M. 2002. GENSTAT Programs for Spatial Analysis of Variety Trials.
ICARDA Biometric Report No. 2, Aleppo, Syria. 38 p.
Smith, A.; B. Cullis and R. Thompson. 2001.
Analysing variety by environment data using multiplicative mixed models and
adjustments for spatial field trend. Biometrics 57: 1138-1147.
Stafford, J. V. 1999. An investigation into the
within-field spatial variability of grain quality. In: J.V. Stafford (Ed).
Proceeding of 2nd European Conference on Precision Agriculture. Sheffield
Academic Press, UK. p. 353-361.
Wilkinson, G. N.; S. R., Eckert; T. W. Hancock and O. Mayo. 1983. Nearest neighbor (NN) analysis of field
experiments. J. of Royal Stat. Soc. 45: 152-212.
Página diseñada por Prof. Jesús Rafael Méndez Natera
TABLA DE CONTENIDO DE LA REVISTA CIENTÍFICA UDO
AGRÍCOLA



